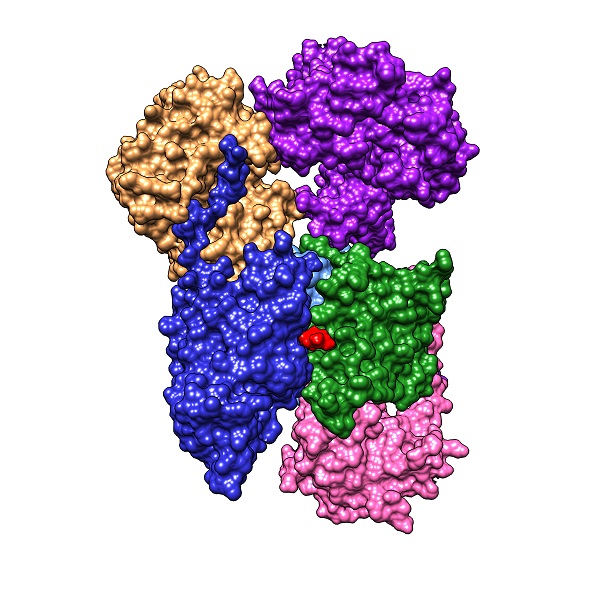
The research groups of Oscar Juarez, assistant professor of biology, and David Minh, assistant professor of chemistry, have discovered a new structural motif in an essential Vibrio cholerae enzyme that could lead to important drug development.
The increasing threat from multi-drug-resistant strains of disease-causing bacteria means that we need to develop novel antibiotics that target essential structures and mechanisms of these microorganisms. The main research focus of Juarez and his group is an essential protein for cell proliferation and pathogenicity of infectious bacterial species such as Vibrio cholerae, Chlamydia trachomatis, Pseudomonas aeruginosa, and numerous others. This enzyme is the sodium-dependent NADH dehydrogenase (Na+-NQR), a cell membrane protein that couples the transport of electrons to the pumping of sodium ions (Na+) across and outside of the membrane, establishing a Na+ gradient that can be used by the bacterium to power essential metabolic processes.
One analogy for Na+-NQR is a watermill. Energy is generated as water flows through the wheel. For Na+-NQR, electrons act as the water, moving within the enzyme through a series of non-protein chemical molecules called cofactors, from a high to subsequent lower energy states, generating energy that the enzyme can use to pump sodium ions. Without the cofactors within the enzyme, electrons could not be transported and thus they are vitally important to the operation of the enzyme.
In a recent scientific article published in the Journal of Biological Chemistry, [1] Juarez’s research group detailed its discovery involving the precise location of the final electron-accepting cofactor, ubiquinone, within the enzyme. A previous publication [2] by the Juarez lab identified HQNO (N-oxo-2-heptyl-4-Hydroxyquinoline), based on enzyme kinetics data, to be an inhibitor competing for the ubiquinone binding site within Na+-NQR, disrupting its function. However, the location of ubiquinone binding site has been highly debated. In collaboration with Minh, computer simulations and molecular docking models were used to guide the functional analysis of different parts of the enzyme. By characterizing the mutants using enzyme kinetics as well as UV-vis spectroscopy, the research team was able to identify the location of the ubiquinone binding site in the interface of subunits B and D of the enzyme. With this information, electron transport within the enzyme and the inhibiting mechanism of ubiquinone analogs such as HQNO can be understood more thoroughly on the structural level, facilitating drug discovery targeting Na+-NQR.
1. Tuz, K. et al. Identification of the Catalytic Ubiquinone-binding Site of Vibrio cholerae Sodium-dependent NADH Dehydrogenase. Journal of Biological Chemistry 292,3039–3048 (2017).
2. Tuz, K. et al. The Kinetic Reaction Mechanism of the Vibrio cholerae Sodium-dependent NADH Dehydrogenase. Journal of Biological Chemistry 290,20009–20021 (2015).